First, some background
Plasma cells are a type of white blood cell made in your bone marrow. Different plasma cells produce different antibodies. There are several subsets (isotypes) of antibodies, and within each subset are more specific kinds of antibodies (made up of different chains of amino acids), each targeting a specific antigen — the toxic molecules that prompt the body's immune response.
Like any cell, plasma cells can become cancerous, resulting in multiple myeloma, which has a five-year survival rate below 50 percent.
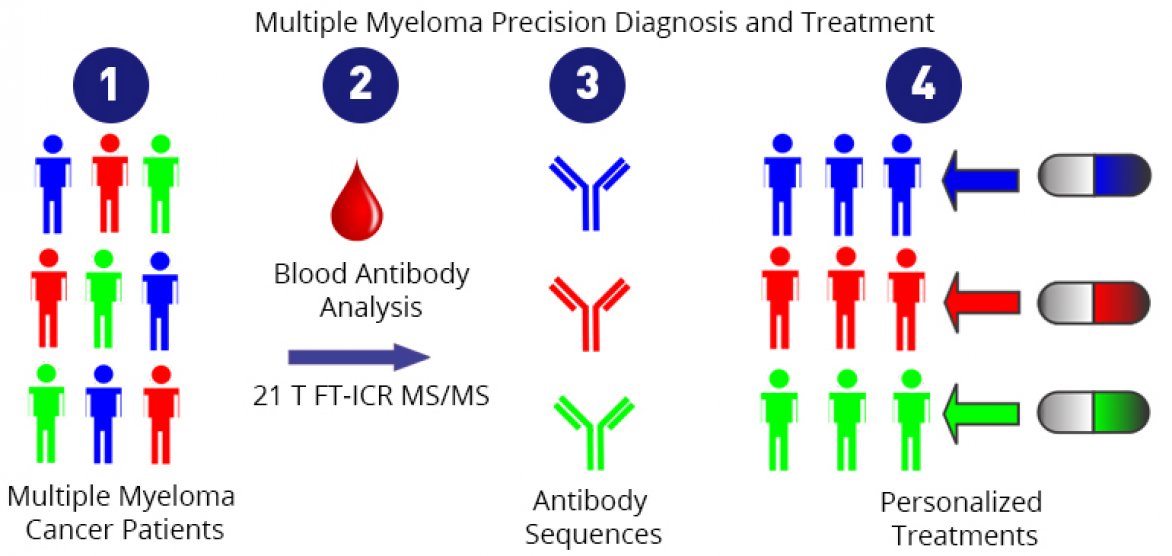
Currently, if a doctor suspects a patient has this disease, she will use a method called gel electrophoresis to test his blood serum and urine for antibodies: elevated levels of a specific antibody (a monoclonal antibody) are a symptom of multiple myeloma.
The same test is used to monitor the effectiveness of treatment, and this is where gel electrophoresis hits a wall. Say a patient experiences a relapse, and tests show an abundance of antibody as seen in the pre-treatment test. While this indicates cancer, it is not clear which antibodies are producing that response, i.e., which plasma cells are cancerous. A different population of plasma cells, producing a different monoclonal antibody, could now be cancerous. It’s also possible that the cancerous cells are the same as those found when the disease was first diagnosed, but that the cancer cells have mutated. It's an important distinction: If a doctor knew which plasma cells were cancerous, she could better target them.
What did scientists discover?
Scientists using instruments at the MagLab's Ion Cyclotron Resonance (ICR) Facility were able to tell that difference, determining the precise molecular makeup of antibodies in a sample. Using a state-of-the-art mass spectrometer powered by world's most powerful ICR magnet (21 teslas), they were able to discern the minute differences in amino acid chains that distinguish one specific antibody from another, effectively identifying biomarkers at the finest level.
In addition, they were able to make these measurements from far lower concentrations of the biomarker (10 times lower) than is doable with present-day conventional methods.
Why is this important?
This new approach could one day enable a molecular level understanding of this type of cancer. In the future, the detailed protein sequencing could lead to precision diagnosis, myeloma cell mutation status monitoring, and personalized cancer treatments. Because of the lower concentration thresholds, physicians may be able to identify the presence of cancer much sooner than previously possible.
Who did the research?
L. He1, L.C. Anderson2, D.R. Barnidge3, D.L. Murray3, C.L. Hendrickson1,2, A.G. Marshall1,2
1Department of Chemistry and Biochemistry, Florida State University; 2Ion Cyclotron Resonance Program, National MagLab; 3Department of Laboratory Medicine and Pathology, Mayo Clinic
Why did they need the MagLab?
The MagLab's 21-tesla ICR mass spectrometer provides the ultra-high mass resolving power, mass accuracy, sensitivity and spectral acquisition rate required to achieve the low detection limit and extensive biomarker sequencing demonstrated in this groundbreaking research.
Details for scientists
- View or download the expert-level Science Highlight, Multiple Myeloma Model Analysis by 21 Tesla FT-ICR Mass Spectrometry
- Read the full-length publication, Analysis of Monoclonal Antibodies in Human Serum as a Model for Clinical Monoclonal Gammopathy by Use of 21 Tesla FT-ICR Top-Down and Middle-Down MS/MS , in Journal of the American Society for Mass Spectrometry
Funding
This research was funded by the following grants: G.S. Boebinger (NSF DMR-1157490)
For more information, contact Chris Hendrickson.